In my last article, I talked about epigenetics and how we can measure epigenetic changes (read here if you missed it). Now we’re moving on to the good stuff—how we assess epigenetic aging and what it means for biological and psychological research.
Many Different Clocks
With about 20,000 protein-coding genes, there is a big question of which genes are important for aging. Scientists have been working toward this answer for a while. Aging is a major process undoubtedly involving many different genes, but it is important to identify the most relevant ones that actually affect aging. Scientists walk a fine line between accuracy and ease. Think of it this way—the more genes we assess, the more accurate our data, but the emptier our wallets. It is important to find easy, and cost-feasible methods to ensure that people can carry out this research in a variety of settings. Dr. Gregory Hannum posited a model involving 71 epigenetic sites (Hannum et al, 2013). This number of sites has greatly ranged, from as many as 589 to as few as 3 (Teschendorff et al, 2010; Weidner et al, 2014). Some models are also specific to certain types of cells (i.e., red blood cells can be assessed). For the bulk of this article, I’ll be talking about a model proposed by Dr. Steve Horvath, a researcher at UCLA, which involves 353 epigenetic sites and is not cell-specific.
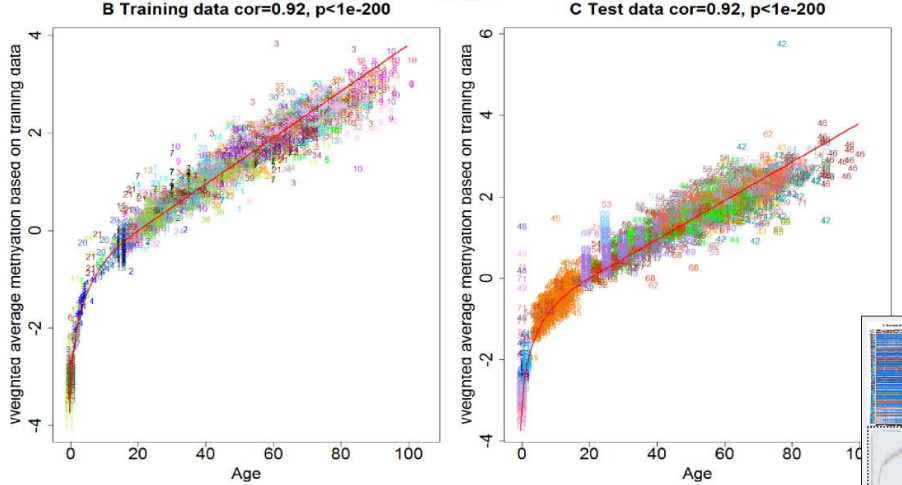
Horvath’s epigenetic clock is renowned for its high accuracy, with a correlation of 0.90 between the epigenetic age and a person’s chronological age. Put another way, over 80% of the variance in epigenetic age is directly related to chronological age. Someone who is 25 years-old likely has cells with an epigenetic age of approximately 25 years-old according to this model, so we can predict chronological age very accurately from epigenetic age. What is most important about Horvath’s model is that age can be calculated from almost any cells in the body with the same formula. Other models would calculate age differently based on the tissue. For example, using another model, you might not derive the same age from your blood cells as your muscle cells, so you would need a different formula to account for the difference. However, there is just one simple calibration curve for Horvath’s model.
Horvath’s calibration curve begins exponentially between embryonic stages and adulthood and then increases linearly (Horvath, 2013).
Health and Research Applications
You might be wondering, “So what? I know my birth day, so why would I need to check my cells for this.” Horvath’s model suggests that all of your cells should be the same chronological age. However, we know that diseases unhealthy behaviors can strain certain parts of the body. Even though HIV can be treated, contraction of HIV is still associated with Not only does smoking damage your lungs and increase risk for lung cancer, it can also increase blood epigenetic aging and alter DNA methylation (Breitling et al., 2011). Thus, a smoker who is chronologically 25 years-old should have cells with an epigenetic age of 25 years-old, but their cells may have an older age because smoking tobacco is associated with more rapid aging. It is worth noting that cancer is a genetic disease, and most cancers naturally occur among the elderly. Using the epigenetic clock, the age of these tissues can be compared with the age of other cells to determine if certain cells are experiencing an accelerated aging and if this aging is influenced by health behaviors.
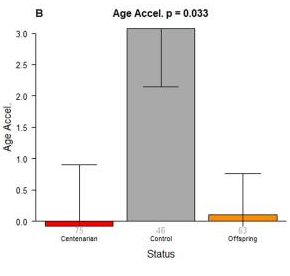
No differences are found between age acceleration in centernarians and their offspring. These data suggest that age acceleration is hereditary, in support of the epigenetic clock (Horvath et al., 2015).
The curve can even answer some of our questions about normal development. The epigenetic clock does not move at a constant rate—methylation increases exponentially until early adulthood, and then levels off to a constant ticking rate for the rest of the lifespan (Horvath, 2013). We may be able to learn more about natural maturation and puberty. For instance, one longitudinal study found that the rate of accelerated aging (the difference between epigenetic age and biological age) at birth predicts greater weight gain and BMI during childhood and adolescence, although accelerated aging during childhood only predicted adolescent height (Simpkins et al., 2017). These results suggest that epigenetic aging is important for development, but its importance varies throughout the lifespan. Also, it’s important to note that the rate of aging is hereditary, and age acceleration may be inherited from your parents. Previous studies suggest that centernarians (people who lived over 100 years) and their children did not differ with respect to age acceleration, but both showed much lower rates of age acceleration relative to people who were not the children of centernarians (Horvath et al., 2015).
So what is driving this natural ticking? What exactly does it represent, and how do we stop it? The short answer is that we still don’t know. However, it seems like the epigenetic clock is related to DNA maintenance. Whenever your cells replicate DNA (which is pretty often), a mistake can occur. A mistake in your DNA can cause serious damage and lead to many different conditions, but thankfully cells have a built-in system for maintaining DNA. There are plenty of proteins involved in identifying and fixing these mistakes. Unfortunately, it seems that the more this system is active, the more aging occurs. This would suggest that we age more rapidly when our cells are trying to repair themselves more. This would make sense in the context of cancer and unhealthy behaviors (Horvath, 2013). For example, if your skin is exposed to UV rays, your cells will be trying to repair the DNA mutations and may consequently age faster.
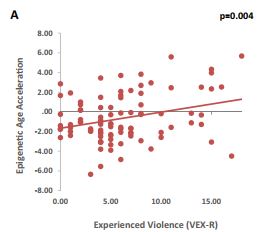
In addition to disease, our biology and our environment can shape the rate of epigenetic aging. Epigenetic processes are a normal part of maturation and, based on growing work with animal models, have been theorized to influence puberty in humans, especially with respect to brain maturation (Morrison, Rodgers, Morgan, & Bale, 2014). One study of children has found that lower socioeconomic status and more experiences of violence both separately predicted greater rates of epigenetic aging; simply witnessing violence was not significantly related to epigenetic age acceleration (Jovanovic et al., 2017). More specifically, they looked at how closely chronological age related to biological age (age measured from their blood using Horvath’s epigenetic clock) for each participant. They took the difference between these ages (i.e., if someone was 10-years-old but their age according to Horvath’s epigenetic clock was 14, they would have an age acceleration of 4). They then found that these rates of age acceleration could be predicted by factors of socioeconomic status (i.e., parental education and household income) as well as experienced violence. Another study found that early parental stress can also predict epigenetic changes in teenagers (Essex et al., 2013). Effects did vary by gender, as maternal stress predicted gene expression in boys and girls whereas paternal stress only predicted gene expression in girls. The results of these studies represent the first steps toward assessing the effects of early stress on aging throughout childhood and adolescence. However, it is important to note limitations within these studies. First, the results regarding SES and violence should still be taken with some caution as these children were specifically samples from a long-term study on trauma in early childhood. Second, note that the study assessing early stress did not use Horvath’s clock (Horvath’s paper wasn’t published when they were running the study). They actually assessed a lot of genes (over 18,000) when assessing links with early stress. Therefore we can’t make a conclusive argument about how adversity affects aging specifically. As measures of epigenetic aging become more widespread, more research can be conducted with larger populations.
More experiences of neighborhood violence predicted greater epigenetic age acceleration, even after accounting for income, parental education, and child sex (Jovanovic et al., 2017).
Epigenetic aging has even been linked with a psychological construct—self-control. One of the first links identified has been between epigenetic aging and self-control. Among African American teens, those exhibiting high self-control at age 17 showed better psychological functioning at age 20 (Miller, Yu, Chen, & Brody, 2015). They had fewer depressive symptoms, internalizing problems, substance use, and aggressive behaviors—all positive outcomes. However, self-control was also related to epigenetic aging, and the same results were found using both Hannum’s and Horvath’s models of the epigenetic clock. Teens of low socioeconomic status showed age acceleration whereas teens of higher socioeconomic status showed age deceleration as self-control increased. Even though these teens are doing better psychologically, they may be experiencing deficits to their health because of limitations in their environment.
The epigenetic clock has been associated with several diseases, including genetic diseases like cancer and HIV. These diseases are directly related to damage to DNA, and thus provide an interesting starting point for applying the epigenetic clock. It has also been linked to other diseases, such as Alzheimer’s, Parkinson’s disease, and Down Syndrome. There is some (very tentative) evidence that epigenetic aging may relate to severity of post-traumatic stress disorder (Boks et al, 2015; Wolf et al, 2016). There is potential that the epigenetic clock can help us to understand how and why these diseases operate.
You might come away from this article thinking ‘okay, a lot of potential—but where’s the data!?’ My answer (although unsatisfying) is that it is only a matter of time. Epigenetic aging is still a novel concept, and there are many people actively using it in their research. Given how long the research process it is, we may see great findings come of this model of aging, or even better models of the clock may come out. The big takeaway is that chronological age is not the whole story, and as more studies of aging come out it’s important to understand what they mean by ‘biological age.’ One of the age-old questions in society is how do we age (and how do we stop it), so it is exciting to see where this new avenue of research will lead the fields of psychology and biology.
References
Boks, M. P., van Mierlo, H. C., Rutten, B. P., Radstake, T. R., De Witte, L., Geuze, E., … & Vermetten, E. (2015). Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology, 51, 506-512.
Breitling, L. P., Yang, R., Korn, B., Burwinkel, B., & Brenner, H. (2011). Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. The American Journal of Human Genetics, 88(4), 450-457.
Essex, M. J., Thomas Boyce, W., Hertzman, C., Lam, L. L., Armstrong, J. M., Neumann, S., & Kobor, M. S. (2013). Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child development, 84(1), 58-75.
Hannum, G., Guinney, J., Zhao, L., Zhang, L., Hughes, G., Sadda, S., … & Deconde, R. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular Cell, 49(2), 359-367. doi: 10.1016/j.molcel.2012.10.016
Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14(10). doi:10.1186/gb-2013-14-10-r115
Horvath, S., Pirazzini, C., Bacalini, M. G., Gentilini, D., Blasio, A. M., Delledonne, M., . . . Franceschi, C. (2015). Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging, 7(12), 1159-1170. doi:10.18632/aging.100861
Jovanovic, T., Vance, L. A., Cross, D., Knight, A. K., Kilaru, V., Michopoulos, V., … & Smith, A. K. (2017). Exposure to violence accelerates epigenetic aging in children. Scientific reports, 7.
Miller, G. E., Yu, T., Chen, E., & Brody, G. H. (2015). Self-control forecasts better psychosocial outcomes but faster epigenetic aging in low-SES youth. Proceedings of the National Academy of Sciences, 112(33), 10325-10330.
Morrison, K. E., Rodgers, A. B., Morgan, C. P., & Bale, T. L. (2014). Epigenetic mechanisms in pubertal brain maturation. Neuroscience, 264, 17-24.
Simpkin, A. J., Howe, L. D., Tilling, K., Gaunt, T. R., Lyttleton, O., McArdle, W. L., … & Relton, C. L. (2017). The epigenetic clock and physical development during childhood and adolescence: longitudinal analysis from a UK birth cohort. International journal of epidemiology, 46(2), 549-558.
Teschendorff, A. E., Menon, U., Gentry-Maharaj, A., Ramus, S. J., Weisenberger, D. J., Shen, H., … & Savage, D. A. (2010). Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome research, 20(4), 440-446. doi: 10.1101/gr.103606.109
Weidner, C. I., Lin, Q., Koch, C. M., Eisele, L., Beier, F., Ziegler, P., … & Zenke, M. (2014). Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biology, 15(2), 1. doi: 10.1186/gb-2014-15-2-r24
Wolf, E. J., Logue, M. W., Hayes, J. P., Sadeh, N., Schichman, S. A., Stone, A., … & Miller, M. W. (2016). Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology, 63, 155-162.